Machine learning algorithms were developed to analyze and predict protein synthesis, enhancing process optimization in various industries. A toolbox and manual were created to facilitate interdisciplinary communication. The research identified key data correlations and variable predictive accuracy for different proteins, revealing that not all influencing factors are captured by the existing data. Future work includes determining optimal data set sizes, improving model quality, and creating interfaces for iterative model improvement and enhanced interpretability.
| Topic Fields | |
| Published | 2022 |
| Involved Institutes | |
| Project Type | ICNAP Research/Transfer Project |
| Result Type | |
| Responsibles |
Proteins are biological macromolecules and a central building block of life on our planet. For use in therapeutics, diagnostics or the food industry, proteins are synthesized in living cells. Due to numerous and partly unknown influencing factors and interactions, these biological processes are highly complex and difficult to predict.
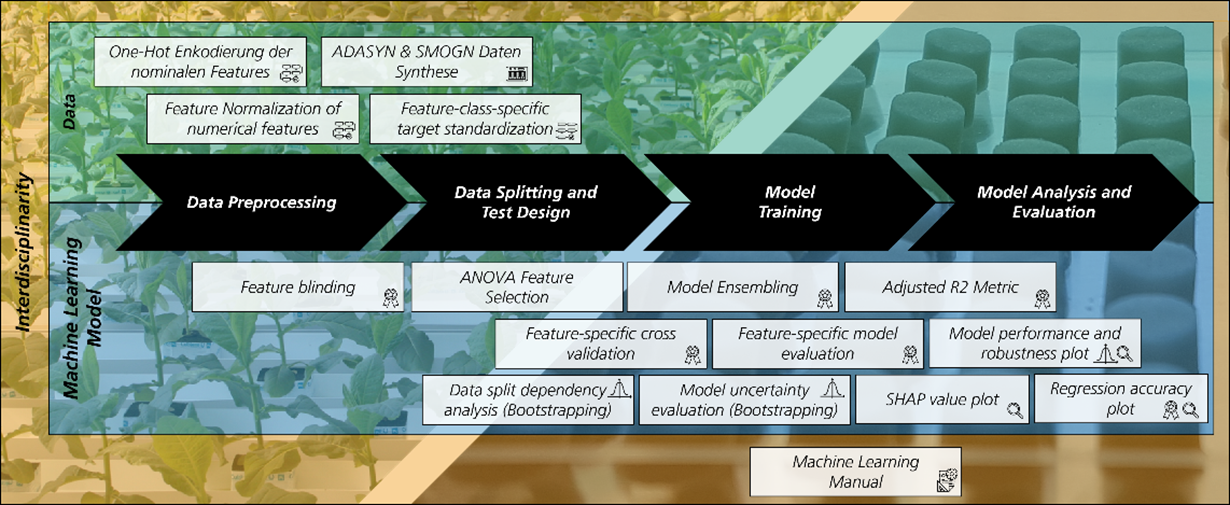
Machine learning algorithms enable the analysis of high-dimensional process data and the creation of predictive quality models and therefore open new possibilities for process optimization. This requires close cooperation between data engineers and domain experts, and a mutual understanding of each other's work, routines, and language. In this project, machine learning algorithms were developed to analyze and model protein synthesis, with a focus on model transparency and comprehensive evaluation.
The developed methods were integrated into a toolbox and a machine learning manual was designed to improve interdisciplinary communication. Insights were gained about data correlations, process influencing factors, and that some proteins can be modeled much more accurately than others. A high variance across multiple predictive models implies that the available data do not capture external factors that significantly affect protein production.
There are several follow-up options to this project, including analyses of the data set size necessary to build reliable models for such variable input data, advanced model quality analysis and an integrated interface between machine learning modelling tools and expert domain knowledge for iterative model improvement and increased interpretability and plausibility.
Contact us to get in touch! With a membership, you’ll gain full access to all project information and updates.
© Fraunhofer 2026